物种分布模型¶
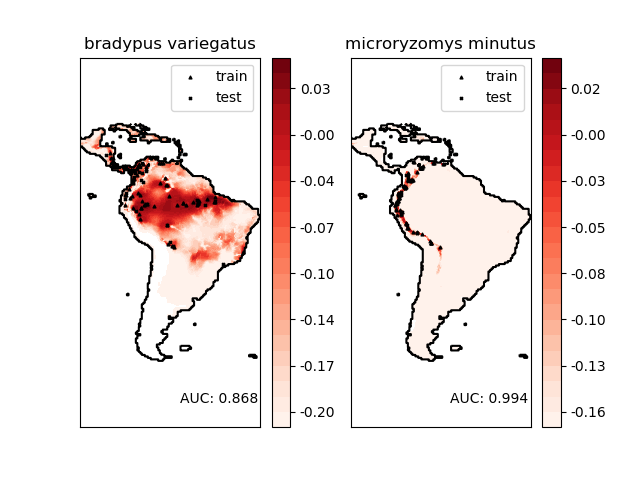
物种地理分布的模拟是保护生物学中的一个重要问题。在这个例子中,我们模拟了两种南美哺乳动物的地理分布,给出了过去的观测结果和14个环境变量。由于我们只有正的例子(没有不成功的观测结果),所以我们把这个问题归结为密度估计问题,并使用 sklearn.svm.OneClassSVM作为我们的建模工具。数据集由Phillips et. al. (2006)提供。如果有,该示例使用basemap绘制南美洲的海岸线和国家边界。
这两个物种是:
“Bradypus variegatus” , the Brown-throated Sloth. “Microryzomys minutus” , 又称森林小稻鼠,一种生活在秘鲁、哥伦比亚、厄瓜多尔、秘鲁和委内瑞拉的啮齿动物。
参靠
“Maximum entropy modeling of species geographic distributions” S. J. Phillips, R. P. Anderson, R. E. Schapire - Ecological Modelling, 190:231-259, 2006.
________________________________________________________________________________
Modeling distribution of species 'bradypus variegatus'
- fit OneClassSVM ... done.
- plot coastlines from coverage
- predict species distribution
Area under the ROC curve : 0.868443
________________________________________________________________________________
Modeling distribution of species 'microryzomys minutus'
- fit OneClassSVM ... done.
- plot coastlines from coverage
- predict species distribution
Area under the ROC curve : 0.993919
time elapsed: 29.37s
# Authors: Peter Prettenhofer <peter.prettenhofer@gmail.com>
# Jake Vanderplas <vanderplas@astro.washington.edu>
#
# License: BSD 3 clause
from time import time
import numpy as np
import matplotlib.pyplot as plt
from sklearn.utils import Bunch
from sklearn.datasets import fetch_species_distributions
from sklearn import svm, metrics
# if basemap is available, we'll use it.
# otherwise, we'll improvise later...
try:
from mpl_toolkits.basemap import Basemap
basemap = True
except ImportError:
basemap = False
print(__doc__)
def construct_grids(batch):
"""Construct the map grid from the batch object
Parameters
----------
batch : Batch object
The object returned by :func:`fetch_species_distributions`
Returns
-------
(xgrid, ygrid) : 1-D arrays
The grid corresponding to the values in batch.coverages
"""
# x,y coordinates for corner cells
xmin = batch.x_left_lower_corner + batch.grid_size
xmax = xmin + (batch.Nx * batch.grid_size)
ymin = batch.y_left_lower_corner + batch.grid_size
ymax = ymin + (batch.Ny * batch.grid_size)
# x coordinates of the grid cells
xgrid = np.arange(xmin, xmax, batch.grid_size)
# y coordinates of the grid cells
ygrid = np.arange(ymin, ymax, batch.grid_size)
return (xgrid, ygrid)
def create_species_bunch(species_name, train, test, coverages, xgrid, ygrid):
"""Create a bunch with information about a particular organism
This will use the test/train record arrays to extract the
data specific to the given species name.
"""
bunch = Bunch(name=' '.join(species_name.split("_")[:2]))
species_name = species_name.encode('ascii')
points = dict(test=test, train=train)
for label, pts in points.items():
# choose points associated with the desired species
pts = pts[pts['species'] == species_name]
bunch['pts_%s' % label] = pts
# determine coverage values for each of the training & testing points
ix = np.searchsorted(xgrid, pts['dd long'])
iy = np.searchsorted(ygrid, pts['dd lat'])
bunch['cov_%s' % label] = coverages[:, -iy, ix].T
return bunch
def plot_species_distribution(species=("bradypus_variegatus_0",
"microryzomys_minutus_0")):
"""
Plot the species distribution.
"""
if len(species) > 2:
print("Note: when more than two species are provided,"
" only the first two will be used")
t0 = time()
# Load the compressed data
data = fetch_species_distributions()
# Set up the data grid
xgrid, ygrid = construct_grids(data)
# The grid in x,y coordinates
X, Y = np.meshgrid(xgrid, ygrid[::-1])
# create a bunch for each species
BV_bunch = create_species_bunch(species[0],
data.train, data.test,
data.coverages, xgrid, ygrid)
MM_bunch = create_species_bunch(species[1],
data.train, data.test,
data.coverages, xgrid, ygrid)
# background points (grid coordinates) for evaluation
np.random.seed(13)
background_points = np.c_[np.random.randint(low=0, high=data.Ny,
size=10000),
np.random.randint(low=0, high=data.Nx,
size=10000)].T
# We'll make use of the fact that coverages[6] has measurements at all
# land points. This will help us decide between land and water.
land_reference = data.coverages[6]
# Fit, predict, and plot for each species.
for i, species in enumerate([BV_bunch, MM_bunch]):
print("_" * 80)
print("Modeling distribution of species '%s'" % species.name)
# Standardize features
mean = species.cov_train.mean(axis=0)
std = species.cov_train.std(axis=0)
train_cover_std = (species.cov_train - mean) / std
# Fit OneClassSVM
print(" - fit OneClassSVM ... ", end='')
clf = svm.OneClassSVM(nu=0.1, kernel="rbf", gamma=0.5)
clf.fit(train_cover_std)
print("done.")
# Plot map of South America
plt.subplot(1, 2, i + 1)
if basemap:
print(" - plot coastlines using basemap")
m = Basemap(projection='cyl', llcrnrlat=Y.min(),
urcrnrlat=Y.max(), llcrnrlon=X.min(),
urcrnrlon=X.max(), resolution='c')
m.drawcoastlines()
m.drawcountries()
else:
print(" - plot coastlines from coverage")
plt.contour(X, Y, land_reference,
levels=[-9998], colors="k",
linestyles="solid")
plt.xticks([])
plt.yticks([])
print(" - predict species distribution")
# Predict species distribution using the training data
Z = np.ones((data.Ny, data.Nx), dtype=np.float64)
# We'll predict only for the land points.
idx = np.where(land_reference > -9999)
coverages_land = data.coverages[:, idx[0], idx[1]].T
pred = clf.decision_function((coverages_land - mean) / std)
Z *= pred.min()
Z[idx[0], idx[1]] = pred
levels = np.linspace(Z.min(), Z.max(), 25)
Z[land_reference == -9999] = -9999
# plot contours of the prediction
plt.contourf(X, Y, Z, levels=levels, cmap=plt.cm.Reds)
plt.colorbar(format='%.2f')
# scatter training/testing points
plt.scatter(species.pts_train['dd long'], species.pts_train['dd lat'],
s=2 ** 2, c='black',
marker='^', label='train')
plt.scatter(species.pts_test['dd long'], species.pts_test['dd lat'],
s=2 ** 2, c='black',
marker='x', label='test')
plt.legend()
plt.title(species.name)
plt.axis('equal')
# Compute AUC with regards to background points
pred_background = Z[background_points[0], background_points[1]]
pred_test = clf.decision_function((species.cov_test - mean) / std)
scores = np.r_[pred_test, pred_background]
y = np.r_[np.ones(pred_test.shape), np.zeros(pred_background.shape)]
fpr, tpr, thresholds = metrics.roc_curve(y, scores)
roc_auc = metrics.auc(fpr, tpr)
plt.text(-35, -70, "AUC: %.3f" % roc_auc, ha="right")
print("\n Area under the ROC curve : %f" % roc_auc)
print("\ntime elapsed: %.2fs" % (time() - t0))
plot_species_distribution()
plt.show()
脚本的总运行时间:(0分29.592秒)
Download Python source code:plot_species_distribution_modeling.py
Download Jupyter notebook:plot_species_distribution_modeling.ipynb