带结构和无结构的凝聚聚类¶
此示例显示了强制设置连接图以捕获数据中的本地结构的效果。这张图就是20个最近邻的图。
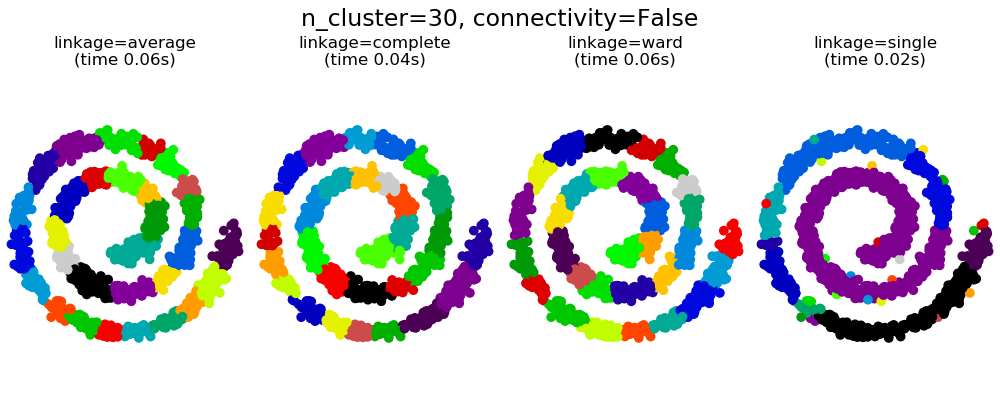
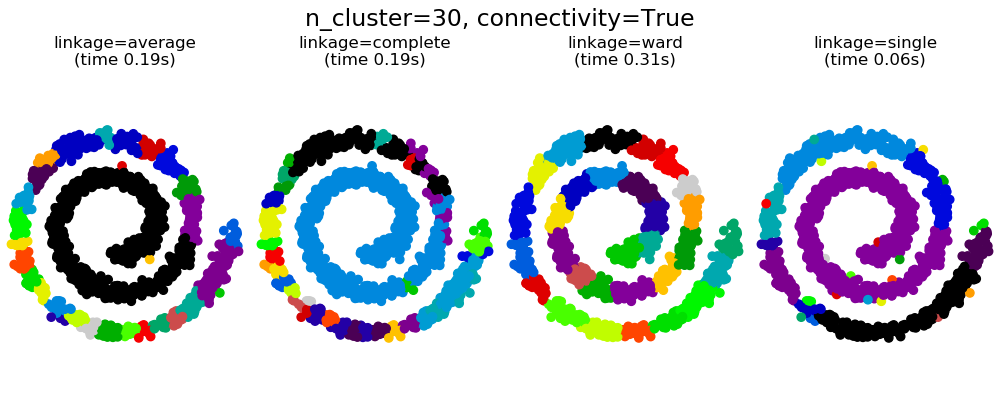
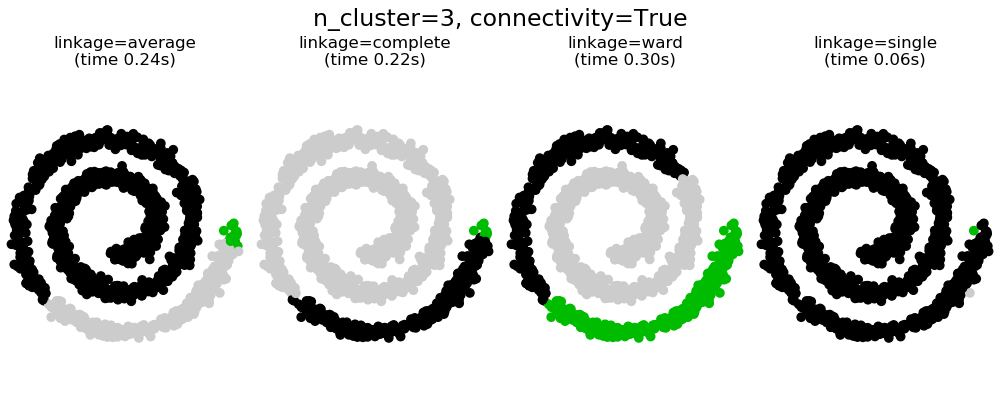
可以看到强制连接的两个结果。第一个有连接矩阵的聚类要快的多。
第二,当使用一个连接矩阵时,单个、平均和完全的联系是不稳定的,并且倾向于创建一些增长非常快的聚类。实际上,平均和完全链接通过在合并时考虑两个聚类之间的所有距离来对抗这种渗流行为(而单个链接只考虑聚类之间的最短距离,从而夸大了它们的行为)。连接图破坏了平均和完整的连接机制,使它们类似于更脆弱的单连杆。这种影响对于非常稀疏的图(尝试减少k-邻域图中的邻居数)和完全链接的效果更为明显。特别是,在图中有非常少的邻居,施加一个接近单一连杆的几何学,这种几何学是众所周知的有这种渗流不稳定性。

# Authors: Gael Varoquaux, Nelle Varoquaux
# License: BSD 3 clause
import time
import matplotlib.pyplot as plt
import numpy as np
from sklearn.cluster import AgglomerativeClustering
from sklearn.neighbors import kneighbors_graph
# Generate sample data
n_samples = 1500
np.random.seed(0)
t = 1.5 * np.pi * (1 + 3 * np.random.rand(1, n_samples))
x = t * np.cos(t)
y = t * np.sin(t)
X = np.concatenate((x, y))
X += .7 * np.random.randn(2, n_samples)
X = X.T
# Create a graph capturing local connectivity. Larger number of neighbors
# will give more homogeneous clusters to the cost of computation
# time. A very large number of neighbors gives more evenly distributed
# cluster sizes, but may not impose the local manifold structure of
# the data
knn_graph = kneighbors_graph(X, 30, include_self=False)
for connectivity in (None, knn_graph):
for n_clusters in (30, 3):
plt.figure(figsize=(10, 4))
for index, linkage in enumerate(('average',
'complete',
'ward',
'single')):
plt.subplot(1, 4, index + 1)
model = AgglomerativeClustering(linkage=linkage,
connectivity=connectivity,
n_clusters=n_clusters)
t0 = time.time()
model.fit(X)
elapsed_time = time.time() - t0
plt.scatter(X[:, 0], X[:, 1], c=model.labels_,
cmap=plt.cm.nipy_spectral)
plt.title('linkage=%s\n(time %.2fs)' % (linkage, elapsed_time),
fontdict=dict(verticalalignment='top'))
plt.axis('equal')
plt.axis('off')
plt.subplots_adjust(bottom=0, top=.83, wspace=0,
left=0, right=1)
plt.suptitle('n_cluster=%i, connectivity=%r' %
(n_clusters, connectivity is not None), size=17)
plt.show()
脚本的总运行时间:(0分3.048秒)
Download Python source code:plot_agglomerative_clustering.py
Download Jupyter notebook:plot_agglomerative_clustering.ipynb