糖尿病数据集上的交叉验证练习¶
使用线性模型交叉验证的教程练习。
本练习在“模型选择”的“交叉验证的估计器”部分中使用:《科学数据处理的统计学习指南》的“选择估计器及其参数”部分。
print(__doc__)
import numpy as np
import matplotlib.pyplot as plt
from sklearn import datasets
from sklearn.linear_model import LassoCV
from sklearn.linear_model import Lasso
from sklearn.model_selection import KFold
from sklearn.model_selection import GridSearchCV
X, y = datasets.load_diabetes(return_X_y=True)
X = X[:150]
y = y[:150]
lasso = Lasso(random_state=0, max_iter=10000)
alphas = np.logspace(-4, -0.5, 30)
tuned_parameters = [{'alpha': alphas}]
n_folds = 5
clf = GridSearchCV(lasso, tuned_parameters, cv=n_folds, refit=False)
clf.fit(X, y)
scores = clf.cv_results_['mean_test_score']
scores_std = clf.cv_results_['std_test_score']
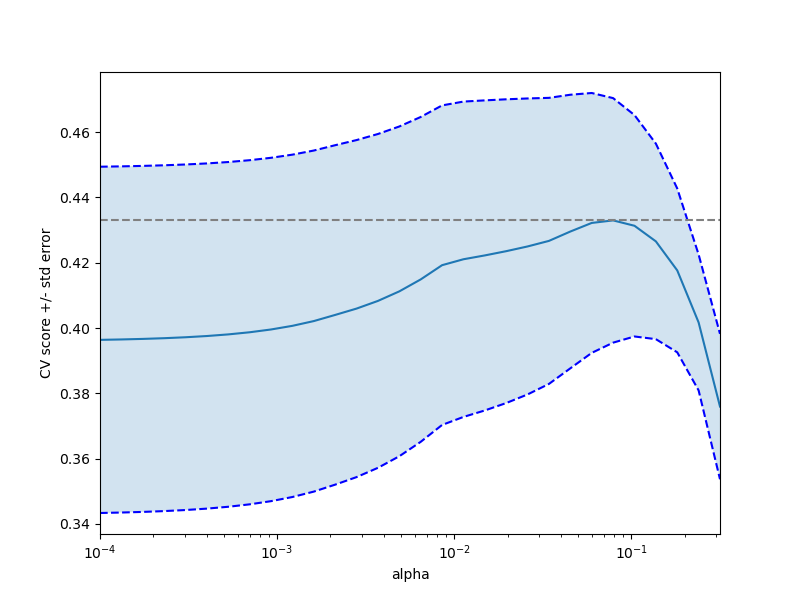
plt.figure().set_size_inches(8, 6)
plt.semilogx(alphas, scores)
# 显示误差线,显示+/-标准。 分数错误
std_error = scores_std / np.sqrt(n_folds)
plt.semilogx(alphas, scores + std_error, 'b--')
plt.semilogx(alphas, scores - std_error, 'b--')
# alpha = 0.2控制填充颜色的半透明性
plt.fill_between(alphas, scores + std_error, scores - std_error, alpha=0.2)
plt.ylabel('CV score +/- std error')
plt.xlabel('alpha')
plt.axhline(np.max(scores), linestyle='--', color='.5')
plt.xlim([alphas[0], alphas[-1]])
# #############################################################################
# 奖励:您对alpha的选择结果有多少信心呢?
'''
为了回答这个问题,我们使用了LassoCV对象,该对象通过内部交叉验证自动从数据中设置其alpha参数(即,它对收到的训练数据执行交叉验证)。
我们使用外部交叉验证来查看自动获得的字母在不同交叉验证折痕之间的差异。
'''
lasso_cv = LassoCV(alphas=alphas, random_state=0, max_iter=10000)
k_fold = KFold(3)
print("Answer to the bonus question:",
"how much can you trust the selection of alpha?")
print()
print("Alpha parameters maximising the generalization score on different")
print("subsets of the data:")
for k, (train, test) in enumerate(k_fold.split(X, y)):
lasso_cv.fit(X[train], y[train])
print("[fold {0}] alpha: {1:.5f}, score: {2:.5f}".
format(k, lasso_cv.alpha_, lasso_cv.score(X[test], y[test])))
print()
print("Answer: Not very much since we obtained different alphas for different")
print("subsets of the data and moreover, the scores for these alphas differ")
print("quite substantially.")
plt.show()
输出:
Answer to the bonus question: how much can you trust the selection of alpha?
Alpha parameters maximising the generalization score on different
subsets of the data:
[fold 0] alpha: 0.05968, score: 0.54209
[fold 1] alpha: 0.04520, score: 0.15523
[fold 2] alpha: 0.07880, score: 0.45193
Answer: Not very much since we obtained different alphas for different
subsets of the data and moreover, the scores for these alphas differ
quite substantially.
脚本的总运行时间:(0分钟0.649 秒)